ActiveAx is a novel approach for producing orientationally-invariant indices of axon diameter and fibre density based on a set of optimized imaging protocols. Axon indices are estimated using the four compartment minimal model of white matter diffusion (MMWMD) described in Alexander et al (2010). Optimized imaging protocols were produced using a experimental framework, a-priori knowledge of the tissue microstructure and scanner hardware as described in Alexander 2008.
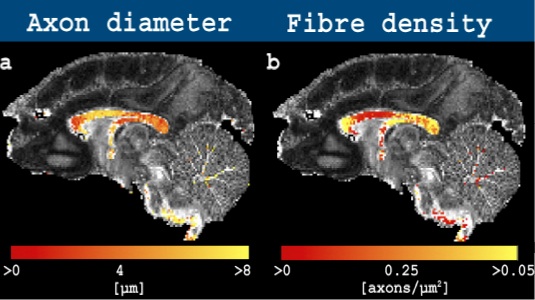
The dataset for download is that used in Alexander et al 2010 and consists of three unique b-values acquired as a high angular resolution diffusion imaging (HARDI) dataset. The dataset includes three slices with a gap. The corpus callosum (CC), a highly-homogeneous bundle of axons, is visible on slice 2 in the slice direction, whereas voxels including more complicated fibre configurations can be found on slices 1 and 3. A tutorial, as well freely-available code to estimate the axon diameter index using AxtiveAx, are presented on the Camino homepage.
Facts about the dataset:
The sequence has been optimized with a total of 360 measurements (N+M) and a Gmax of 140 mT/m and uses a diffusion weighted pulse gradient spin echo (PGSE) with single line readout. Common for the b-values are the image matrix: 265x128, voxel size: 0.4x0.4x0.4 mm^3, slices: 3, TR= 5000 ms, TE= 60 ms, gap = 2 mm and N = 90 non-collinear diffusion weighted directions on the unit-shell available in Camino. For the M=3 unique b-values: Diffusion-weighted gradient strength (Gmax)= [140, 140, 131, 140] mT/m, small delta=[10.2, 10.2, 7.6, 17.7] ms, big delta=[16.7, 16.7, 45.9, 35.8] ms, b-value of [1925, 1931, 3091, 13183] s/mm^2. The small difference between the two lowest b-values is due to round-off errors.
To cite:
Please cite the following, describing how the data was setup and acquired:
Dyrby TB, Baaré WF, Alexander DC, Jelsing J, Garde E, Søgaard LV, An ex vivo imaging pipeline for producing high-quality and high-resolution diffusion weighted imaging datasets, Human Brain mapping, in-press, April, 2011
Please cite the following describing the ActiveAx setup:
Alexander DC, Hubbard PL, Hall MG, Moore EA, Ptito M, Parker GJ, Dyrby TB. Orientationally invariant indices of axon diameter and density from diffusion MRI Neuroimage. 2010 :1374-89
Download the ActiveAx ex vivo dataset here (White Matter Microscopy Database at Open Science Framework)
Download the Camino gradient scheme here
Observed problems or artifacts: None
Funding:
Lundbeck Foundation and The EU founded CONNECT consortium under the FET program


