Welcome to the dataset used in Andersson et al. 2021: “Does powder averaging remove dispersion bias in diffusion MRI diameter estimates within real 3D axonal architectures?”
Aim
This dataset was used to extract and analyze the 3D morphologies of axons in a crossing fiber region of the vervet monkey brain. Diffusion – and a pulsed-gradient spin echo (PGSE) magnetic resonance imaging (MRI) sequence – was then simulated within the segmented axons. This allowed for a validation of powder averaging approaches to axon diameter estimation in diffusion MRI.
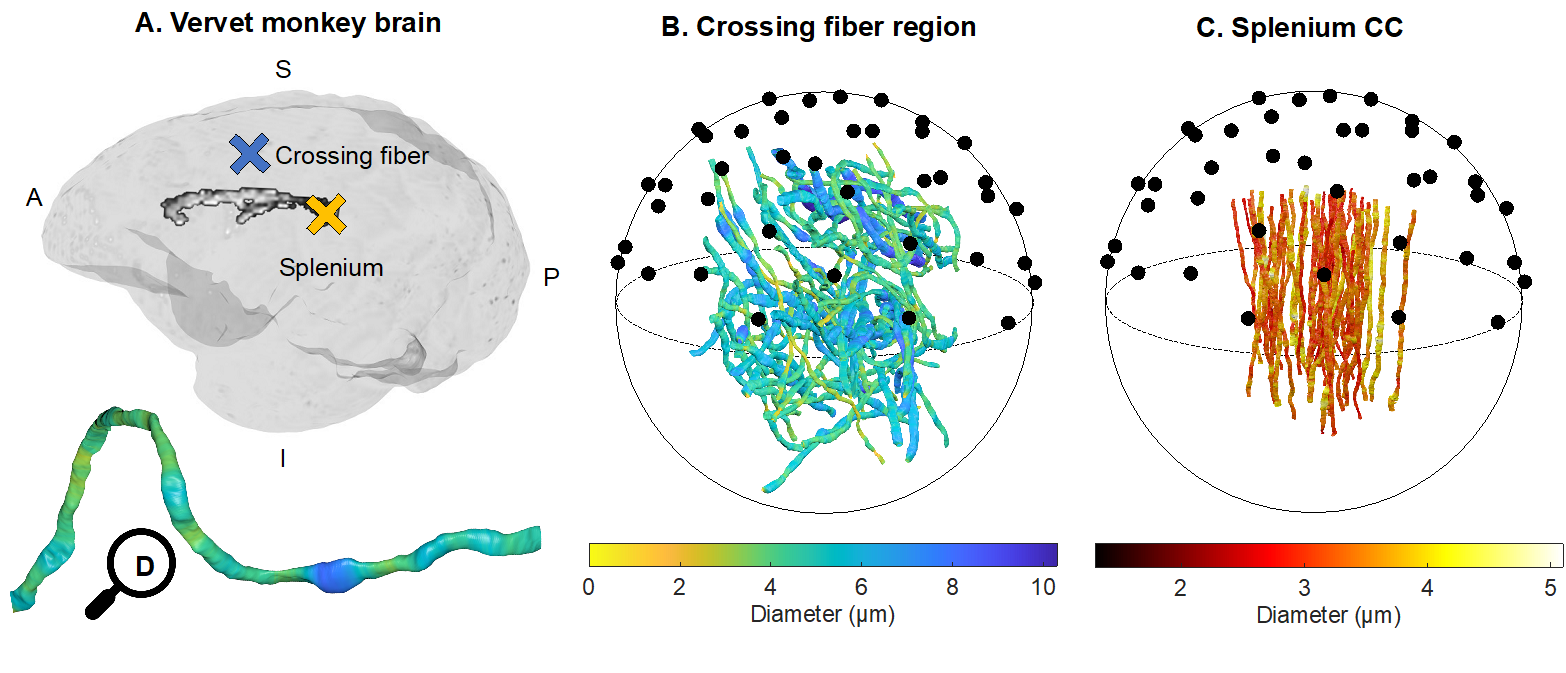
Fig. 1 from Andersson et al. 2021. Can axon diameter be accurately estimated in different WM regions using PA approaches, despite complex axonal morphologies? Validation of PA-estimates of axon diameter was carried out in two different fibre architectures of the vervet monkey brain. A) Axons were segmented from XNH volumes of a heterogeneous crossing fibre region in the anterior semiovale (blue cross) and the splenium (yellow cross). B) 58 segmented crossing fibre axons and C) 54 segmented splenium axons from Andersson et al. (2020). The axon diameters are indicated by the respective colorbars and the black circles represent uniformly distributed directions on the unit sphere. D) A magnified view of one of the axons from B, showing its trajectory and diameter variations.
Data
The data can be downloaded from the following link:
https://resources.drcmr.dk/DIGdata/MAX4Img_PowderAveraging/
The data package here includes a synchrotron X-ray Nano-Holotomography (XNH) volume from a sample taken from a crossing fibre region of a 32-month old vervet monkey, as well as the corresponding segmentations of 58 axons. The XNH volumes from the vervet monkey splenium can be found on https://www.drcmr.dk/axon-morphology-dataset. Additionally, we share the scripts used to estimate axon diameter from simulations of diffusion (and the diffusion MRI signal) within the segmented axons.
In the different directories, the user will find "README.html" files further explaining the data and its details.
The data package is divided into two main sections: “CrossingFibre_XNH_Data” and “Scripts”.
CrossingFibre_XNH_Data
This directory includes the sub-directories:
1) "CrossingFibre_Intensity_voxSize_100nm"
This directory contains the raw, 100 nm isotropic voxel size, X-ray Nano-Holotomography volume of the monkey brain crossing fibre region.
2) " CrossingFibre_Segmentation_voxSize_200nm"
This directory contains a downsampled (by a factor of 2) version of the X-ray Nano-Holotomography volume of the monkey brain crossing fibre region, and the corresponding segmentation of 58 axons.
“Scripts”
This directory contains the scripts needed to fit axon diameter to the diffusion MRI signal (arising only from the intra-axonal space) using the SMT-1, SMT-2 and SMT-3 implementations described in the article.
Methods
Further details can be found in the "Methods" section of the article. In case of questions, please contact one of the corresponding authors, and we will be happy to assist you.
Corresponding authors
Mariam Andersson () and Tim B. Dyrby ()
To cite
Please make sure to cite the listed articles if using or mentioning the following:
- XNH data: Andersson et al. 2020 and Andersson et al. 2021.
- Diameter estimation scripts: Andersson et al. 2021
Funding
The Capital Region Research Foundation and the Swedish Research Council.
References
Mariam Andersson, Marco Pizzolato, Hans Martin Kjer, Katrine Forum Skodborg, Henrik Lundell, Tim B. Dyrby, Does powder averaging remove dispersion bias in diffusion MRI diameter estimates within real 3D axonal architectures?, NeuroImage, 2021, 118718, ISSN 1053-8119.
Mariam Andersson, Hans Martin Kjer, Jonathan Rafael-Patino, Alexandra Pacureanu, Bente Pakkenberg, Jean-Philippe Thiran, Maurice Ptito, Martin Bech, Anders Bjorholm Dahl, Vedrana Andersen Dahl, Tim B. Dyrby. Axon morphology is modulated by the local environment and impacts the noninvasive investigation of its structure–function relationship. Proceedings of the National Academy of Sciences Dec 2020, 117 (52) 33649-33659; DOI: 10.1073/pnas.2012533117


